Getting Started
DSMNC is a database which provides information about somatic mutations (based on UCSC hg19, mm9) in human and mouse normal cells. The database interface supports the capability of browsing
Browse, searching
Search (gene name or genome position) and downloading these somatic point mutations of normal cells. User can download all the data and do more deep analysis
Download.
We recommended to use Google Chrome for the database browsing. Please
Contact Us with any questions.
Filter Data
Select the “SNVs Table” in the “Browse” toolbar menu to browse SNVs tables(Figure 1A). These tables can be narrowed and filtered by selecting one or more values in a category on the left hand side of the page(Figure 1B).
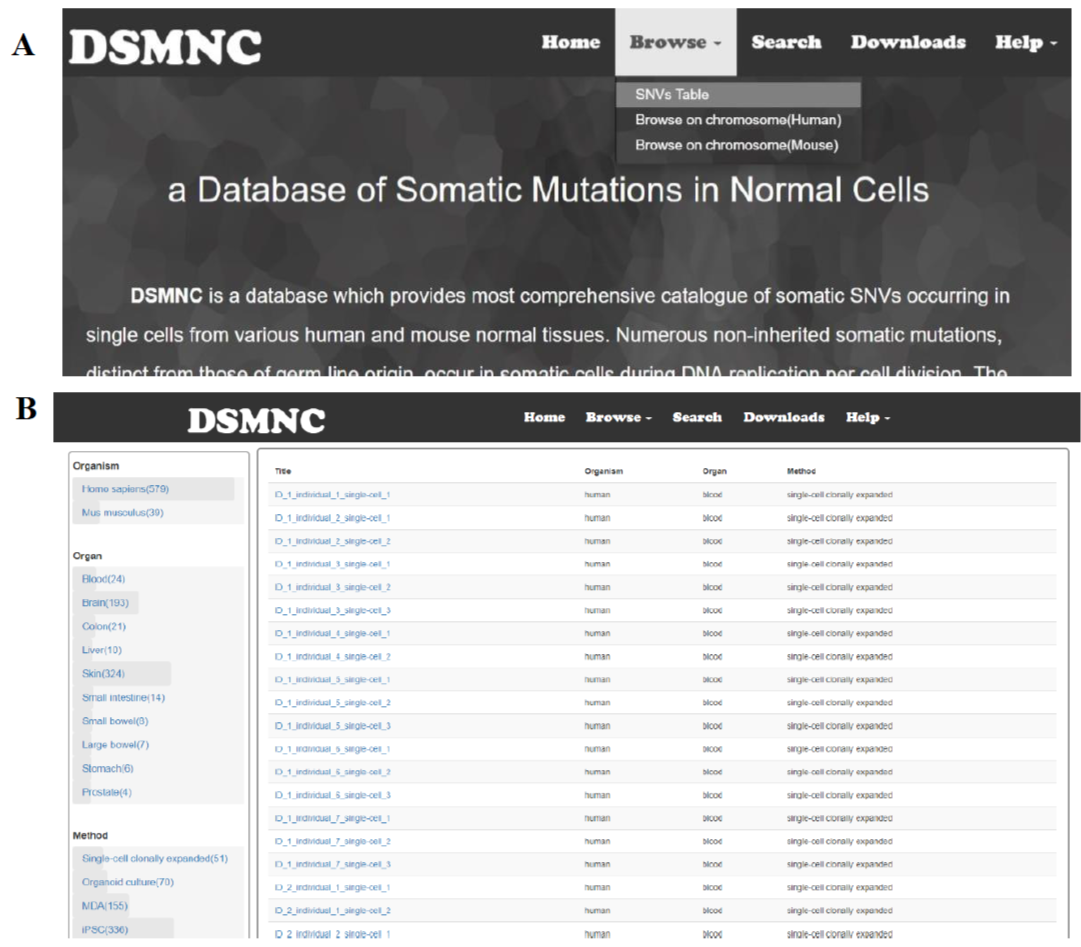
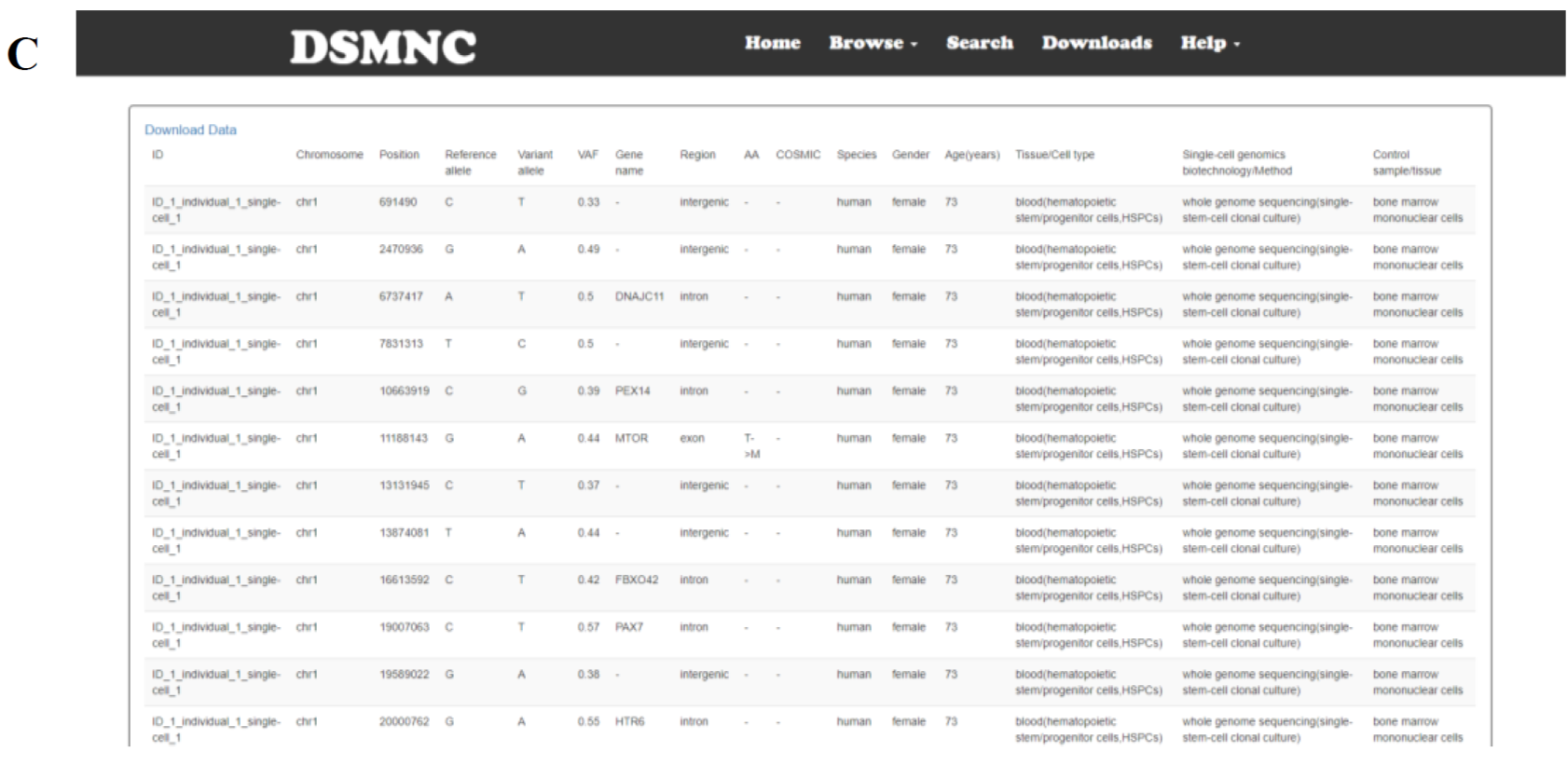
Figure 1. (A) Select the “SNVs Table” in the “Browse” toolbar menu to browse SNVs tables.(B) These tables can be narrowed and filtered by selecting one or more values in a category on the left hand side of the page.
Browse data
Select the “Browse on chromosome(Human)” or “Browse on chromosome(Mouse)”(Figure 2A) in the “Browse” toolbar menu to browse SNVs on JBrowse(Figure 2B).
Moving
·Move the view by clicking and dragging in the track area, or by clicking ←or → in the navigation bar, or by pressing the left and right arrow keys.
·Center the view at a point by clicking on either the track scale bar or overview bar, or by shift-clicking in the track area.
Zooming
·Zoom in and out by clicking or in the navigation bar, or by pressing the up and down arrow keys while holding down "shift".
·Select a region and zoom to it ("rubber-band" zoom) by clicking and dragging in the overview or track scale bar, or shift-clicking and dragging in the track area.
Showing Tracks
·Turn a track on by dragging its track label from the "Available Tracks" area into the genome area, or double-clicking it.
·Turn a track off by dragging its track label from the genome area back into the "Available Tracks" area.
Searching
·Jump to a feature or reference sequence by typing its name in the location box and pressing Enter.
·Jump to a specific region by typing the region into the location box as: chr:start-end.
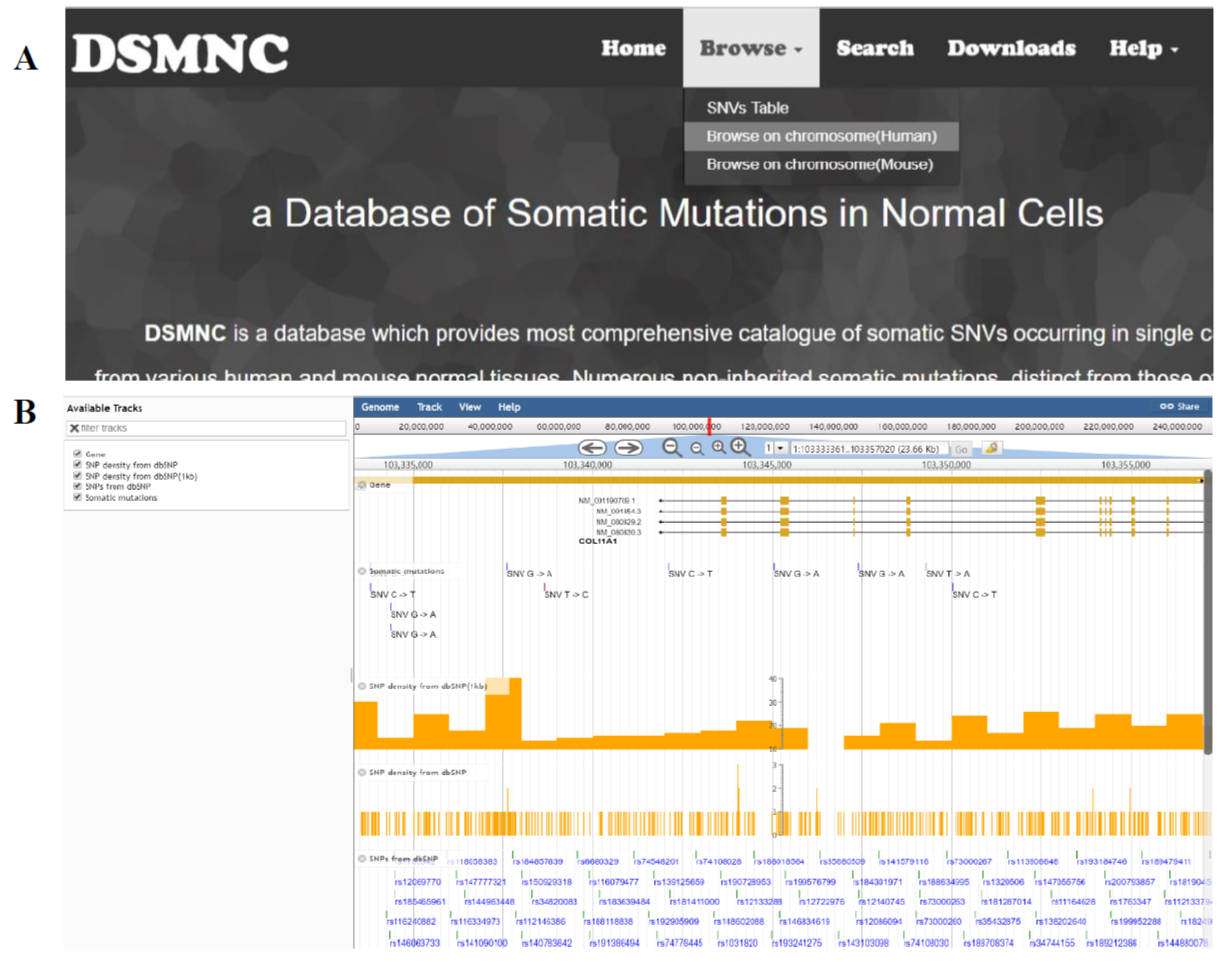
Figure 2. Visualize the somatic mutation data.
Search for data
Select “Search” option in the toolbar. The SNVs data can be searched by entering a searching term in the search box. Searching terms include a region or a gene or gene list(Figure 3). Search a specific region by typing the region into the location box as: chr:start-end and statistics information is also provided in the searching result including somatic mutation frequency, mutation type (C->T, G->A, ...) fraction and synonymous/nonsynonymous mutation ratio within an user specified genomic region. Search by gene is also designed for fuzzy search by choosing the "fuzzy search" button.
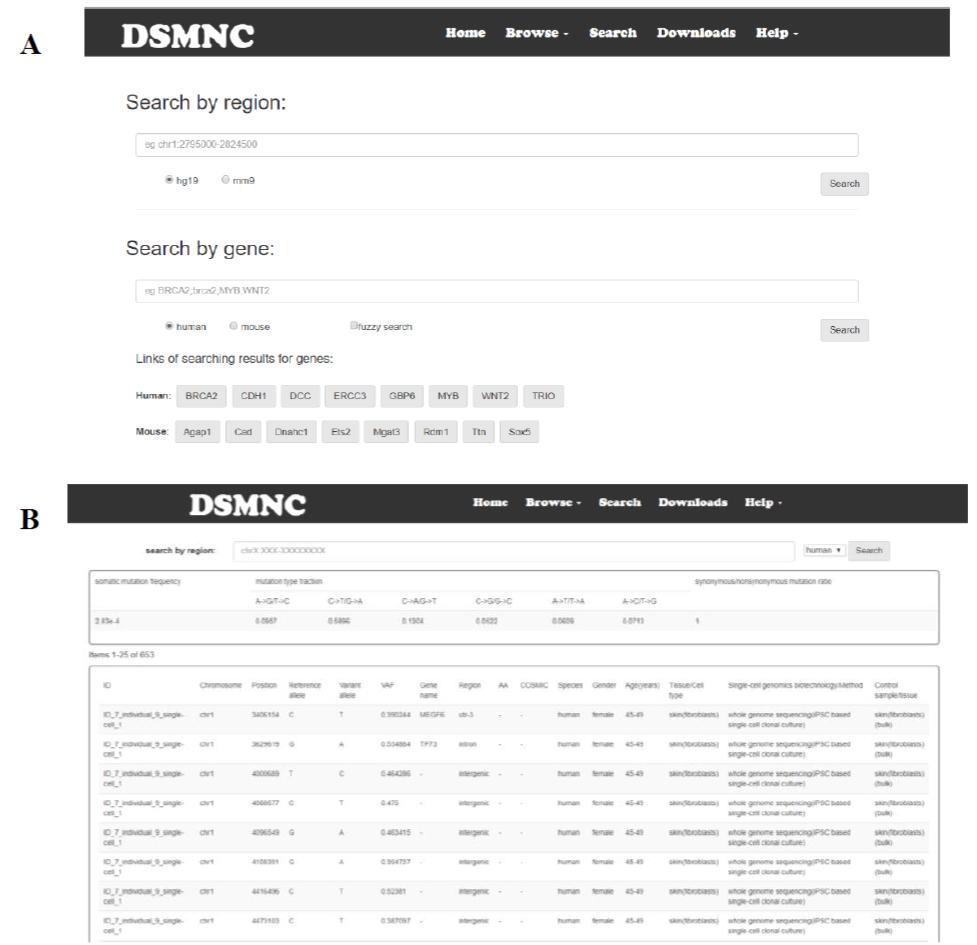
Figure 3. The SNVs data can be searched by entering a searching term in the search box. Searching terms include a region or a gene.